This paper is a final project from the course I teach at the NIH Foundation for Advanced Education in the Sciences — TECH 566: Biotechnology Management
Sandip De
Despite the variety of health systems across hundreds of different countries, one feature is near‐universal: We all depend on private industry to commercialize and market drug products. And because drugs are such an integral part of our health care system, that industry is generally heavily regulated. Yet despite this regulation, little is publicly known about drug development costs. But aggregate research and development (R&D) data are available, and the pharmaceutical industry spends billions per year. The process of developing a new drug to treat a disease or illness is generally long, expensive and full of uncertainty. Several studies have made their effort to estimate the total cost. According to PhRMA (Pharmaceutical Research and Manufacturers of America) in 2010, despite the challenging economic environment, biopharmaceutical companies have invested an estimated $67.4 billion in the search for new medicines of which PhRMA member has invested an estimated $45.8 billion. The trend of expenditure on drug development versus
new drug approval is not correlated since 2000 (Fig. 1)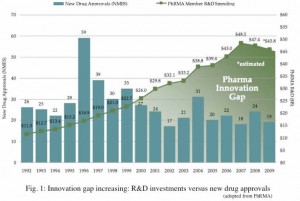 .
.
Pharmaceutical and biopharmaceutical companies need to find several ways to screen for potential problems with promising molecules at the earliest possible stage. They also need to standardize the entire drug development process in such a way that compounds that pass through the initial screening process pass quickly along the developmental pipeline.
About 1‐1.5 % of the newly developed drugs are targeted to cure tropical diseases (1). Commercial pharmaceuticals and patented drug development processes are mainly targeting diseases either prevalent in West or diseases having an impact in the whole world. As the commercial drug development is a costly process, managing financial support to cover research and development for tropical diseases has always been a tough task leaving these diseases unchallenged. Three main proposals have been suggested to solve this problem. The first is providing subsidy by sponsors (government and charities) to the purchases by the developing countries and the second is establishing nonprofit virtual pharmas looking for drug candidates which will go under quick development processes through contract research organizations and corporate partners. The concept of third proposal is to establish a web‐based, community‐wide, decentralized system where scientists and students from universities, institutes and commercial corporations working together to discover drugs against common diseases of interest which is also referred as open source research and development (2).
Generically, open source refers to a program in which the source code is available to the general public for use and/or modification from its original design free of charge. Typically open source code is created as a collaborative effort in which programmers improve upon the code and share the changes within the community.
Open‐source research first started in the software industry around 20 years back and it has become a business model. The best‐known product of this open source culture is Linux, an excellent alternative operating system to Microsoft Windows. The successful sequencing of the genomes of human and several pathogens and non‐pathogenic model organisms has led the way to genomics based methods for drug candidate and drug target prediction, and biology has become an information oriented science.
Open source research and development is a novel approach to research that lets scientists join hands freely across organizations, disciplines and borders to solve problems in which they share an interest. Hollingsworth has already showed that creativity and innovation frequently comes out when diverse brains interact in an unstructured manner (3, 4). According to the open source drug discovery model, there will be a website where volunteers will submit new different projects by using a variety of computer programs and databases searching for new chemicals to attack known biological targets, finding protein targets and posting data from related chemical and biological experiments. The candidates and targets are validated eventually by a set of researchers from public‐private partnership consortium. Open source’s main target is harnessing cheap but precious creativity arising by drawing talents from around the world. These ideas are generally beyond the reach of scientists working in the high‐tech labs either in academia or in pharmaceutical industry. But it is difficult to adopt the open source model in drug discovery research. The research dynamics between software industry and drug development industry significantly differ, development of software only requires computers and internet connections but with drug research these days you not only have to have computers with internet connections but also a financial source to support for laboratory expenses and clinical trials (5). Software development does not involve discovery phase, once the need is identified programmers work steadily to reach their goal and the developmental process is lightly regulated, the quality standards required to release a software once developed is less troublesome. On the other hand drug discovery cannot happen until a certain extent of information is known about the disease, possible candidate drug and it’s target; to collect this information it might take several years to several decades. Drug development
involve coordinated and balanced participation of several disciplines which is very tricky, a little mistake in earlier step can become a much larger problem in later stages leading to utter failure of the whole drug discovery program. To get the FDA approval, a new candidate drug has to undergo rigorous trials, which are strictly regulated and is cumbersome process. Finally, software and drug development industry follow different intellectual property regimes. Software is protected by copyrights where as drug research is patent protected which are costly to file and
maintain.
Despite differences mentioned earlier open source idea has been applied to biological research. The first attempts were in software development in bioinformatics for example Biojava, Bioperl, Biopython etc. Researchers now‐a‐days identify promising drug targets and several prominent lead chemicals using bioinformatic programs. For example, a SARS protein, which is similar to mRNA capmethyltransferase, was identified by comparing proteins’ structures encoded by SARS genome with known protein structures available in the databases. The cap‐methyltransferase has already known inhibitors; so development of a drug similar to the known inhibitors will not be a tough task to eradicate SARS.
There are two best‐known examples where open source drug discovery model has been applied; first one is Medicines for Malaria Venture (MMV), which was established in 1999 to discover new anti‐malarial drugs. An expert scientific advisory committee reviewed projects submitted by volunteers in MMV; the best promising projects are funded and managed by a project manager who outsources the research and development to a network of 260 partners in almost 50 countries. The scientific advisory committee reviews the results after finishing each step of the project to decide whether to continue the project or not. Philanthropic and public partners provide funding of the MMV project. In its search for new molecules against malaria, MMV and partners have so far supported the screening of more than 5 million compounds for their potential activity against the malaria parasite. MMV’s innovative antimalarial pipeline contains over 65 cutting‐edge projects and targets the eradication of malaria. MMV supported the development and final launch in 2009, of Coartem® Dispersible, the first affordable, high quality paediatric ACT for use in Africa.
More recently Tuberculosis disease has been considered as the field for open source drug research and development. This initiative was launched on 15th September 2008. The OSDD‐TB portal has around 146 ongoing projects, some of which addresses complex scientific challenges on drug discovery for tuberculosis. There are about 5390 registered participants from nearly 130 countries putting their effort jointly. The results of the projects are posted in real‐time on the portal in an open electronic lab notebook, which is for community‐wide review. Government of India presently provides the major share of funding, apart from that several philanthropic organizations have contributed to this project. The OSDD community has identified more than 60 potential drug targets of Mycobacterium tuberculosis. Seven targets are being actively followed‐up for onward investigation.
A common argument is that the concept of collaborative drug development has always been active but it has been silent and invisible. The requirement of open source drug discovery was felt in the late 1990s and initiative on open source research and development had taken place quite a while back, but its success has little reached beyond the in silico experiments. Most open source activities occur at a pre-commercial research stage. One main reason people identify is incomplete involvement of researchers due to lack of proper incentives, which they get from traditional patent oriented drug discovery but over the past decade legal scholars, economists, businessman and other social scientists have learned a great deal about the incentives that drive ordinary open source research and development. Researchers are becoming more aware of the positive side of sharing knowledge rather keeping it within, much of the interest in open innovation is directed towards discussing scientific enigmas, not toward discussing commercial projects (6).
In the late 1990s, the idea of open source drug discovery seemed so inevitable that there was no particular need to ask how it would work. In 2004, the US Food and Drug Administration (FDA) launched the Critical Path Initiative to call attention to an alarming decline in the number of innovative medical products being submitted for FDA approval and emphasized that improving R&D productivity required faster incorporation of new
science and improving R&D accuracy (7). On 2006 FDA’s Commissioner announced the release of FDA’s Critical Path Opportunities List containing 76 examples where new scientific discoveries in genomics, bioinformatics and bio-imaging etc. could be used during drug development process to improve the predictability of tests required for assessing the safety and efficacy of potential medical products (7). On 2005 with the mission of improving public health through biomedical research National Institutes of Health (NIH) has established the program on public-private partnerships (PPPs). Perhaps open source is the first fundamentally new innovation mechanism since patents and copyright appeared four centuries ago. Successful collaborative drug discoveries would show that open source methods are not limited to software and could even open the door to applications beyond biology (8). The Pink Army, CollabRX, the African Network for Drug and Diagnostics like new research models are using open source concept for designing customized cancer therapies and new medicines for neglected diseases. In the
past 4-5 years the acceptance of open sourced research and development has become more acceptable by big pharmaceutical companies. Open source research and development is neither inevitable nor impossible, but merely difficult. This paper has intended to show that open source drug discovery does not put the present commercial drug development business at risk but is an area to put more ideas and creativity for generating avenues to resolve the drug discovery crunches.
References:
1. Trouiller O, Olliaro PL Drug development output from 1975 to 1996: What proportion for tropical diseases? International Journal of Infectious Diseases 3: 61–63, 1999.
2. Maurer SM, Rai A, Sali A Finding Cures for Tropical Diseases: Is Open Source an Answer? PLoS Med 1(3): e56, 2004.
3. Hollingsworth JR & Hollingsworth EJ Practicing Interdisciplinarity (eds Weingart P & Stehr N) 215–244 (Univ. Toronto Press, Toronto, 2000).
4. Hollingsworth, J. R. Creating a Tradition of Biomedical Research (ed. Stapleton D) 17–63 (Rockefeller University Press, New York, 2004).
5. Munos B Can open-source R&D reinvigorate drug research? Nature Reviews Drug Discovery 5: 723-729, 2006.
6. Munos B Can Open-Source Drug R&D Repower Pharmaceutical Innovation? Commentaries Clinical Pharmacology & Therapeutics 87(5): 534–536, 2010.
7. Woodcock J, Woosley R The FDA critical path initiative and its influence on new drug development. Annual Reviews Medicine 59: 1-12, 2008.
8. Maurer SM Open source drug discovery: finding a niche (or maybe several) UKMC Law Review 76: 405–435, 2008.